Sixty years ago this month Nature published the famous paper by Watson and Crick solving the structure of DNA. At the time many researchers pursued this goal, made difficult by the complexity of the DNA itself. A key contribution to the solution of the puzzle was the x-ray diffraction data provided by Rosalind Franklin. Indeed, without x-ray diffraction experiments this discovery would have been almost impossible at the time.
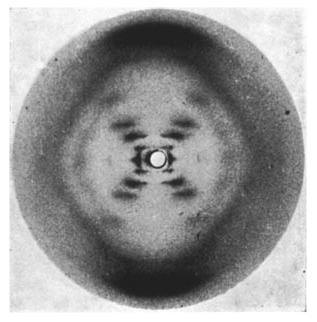
Rosalind Franklin’s x-ray diffraction image of DNA. (c) Nature Magazine. Franklin, R. & Gosling, R. G. Nature 171, 740-741 (1953) – doi:10.1038/171740a0
The way x-ray crystallography works is that a beam of x-rays is directed at a crystal, where the x-rays bounce off the atoms. Because the atoms in a crystal are periodically arranged, the x-rays form complex but regular patterns (such as the one seen for DNA). A detailed analysis of these patterns enables the precise determination of the crystal structure.
To this day such experiments aren’t easy. They require relatively large crystals and typically are done at major facilities such as electron synchrotrons. The synthesis of the crystals for these experiments can often be very difficult.
Yasuhide Inokuma, Makoto Fujita and colleagues from the University of Tokyo in Japan and the University of Jyväskylä in Finland have now developed a clever method that does away with many limitations of x-ray crystallography. Their method works with tiny amounts of material, only about a half to 5 micrograms are enough. This is around a millionth of a gram – truly tiny. The difference between a microgram and a gram is the same as that between a gram and a metric ton. In addition, another major advance of their method is that the target molecules don’t even need to be in a crystalline state. How does this work?

A molecular framework that has absorbed absorbed target molecules (yellow) into a regular array. Reprinted by permission from Macmillan Publishers Ltd. Inokuma, Y. et al. Nature 495, 461–466 (28 March 2013). doi:10.1038/nature11990
The trick used here to avoid all cumbersome crystal synthesis is to simply embed the molecules into a larger host crystal. Metal-organic frameworks (MOF) are particularly suited for this. These are porous three-dimensional crystal structures whose pores and channels can be used to absorb other molecules. In a way, they are like sponges absorbing water. The difference to sponges is only that the MOFs are highly regular themselves. So any target molecule absorbed is held in very regular places as well.
This periodic array of target molecules within the MOF turns out to be just fine for x-ray crystallography. It doesn’t make a difference whether a perfect crystal of only target molecules is synthesised, as has been done in past experiments, or whether as done here they are held in place by embedding them into another crystal – the crystallography works just the same. It is no problem to separate the x-ray signature of the MOF from the target molecule.
This approach has a few advantages. As the authors write themselves in the paper: “Our method solves the real and intrinsic problems of X-ray crystallography and transforms it into a rapid and convenient method for the analysis of molecular structures using only a trace amount of sample.”
Despite this very nice demonstration, there still may be some issues that may still need to be solving. For once, it will be interesting to see how complex and large the measured target molecules can be. And then of course, does the arrangement of the molecules in the MOF really allow for a ultra-precise structure determination? But such questions apart, this new method could fundamentally alter the way we do x-ray spectroscopy.
References:
1. WATSON, J., & CRICK, F. (1953). Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid Nature, 171 (4356), 737-738 DOI: 10.1038/171737a0
2. FRANKLIN, R., & GOSLING, R. (1953). Molecular Configuration in Sodium Thymonucleate Nature, 171 (4356), 740-741 DOI: 10.1038/171740a0
3. Inokuma, Y., Yoshioka, S., Ariyoshi, J., Arai, T., Hitora, Y., Takada, K., Matsunaga, S., Rissanen, K., & Fujita, M. (2013). X-ray analysis on the nanogram to microgram scale using porous complexes Nature, 495 (7442), 461-466 DOI: 10.1038/nature11990


April 9, 2013 at 12:27
Dear Joerg,
Its quite a coincidence that I read this and the article in Nature this week about Crick’s medal and a letter that he wrote to his 12 yr old son with the first ever documented ‘hand-drawn’ structure of DNA! Its fascinating how far we’ve come from that day 🙂
Keep posting ….
SR